| Original research | Peer reviewed |
Cite as: Gamboa-Marín A, Mejía-Wagner DC, Moreno-Ocampo PA, et al. Antimicrobial susceptibility of Listeria monocytogenes, Listeria ivanovii, and Listeria species isolated from swine processing facilities in Colombia. J Swine Health Prod. 2013;21(1):10–21.
Also available as a PDF.
SummaryObjective: To analyze distribution of Listeria monocytogenes serotypes and antimicrobial susceptibility of Listeria isolates from a domestic swine processing facility. Materials and methods: Presumptive Listeria isolates (314) were molecularly identified to discriminate among L monocytogenes, Listeria ivanovii, and Listeria species. Listeria monocytogenes serotypes were identified by polymerase chain reaction (PCR) and PCR-restriction enzyme analysis (PCR-REA) and tested for antimicrobial susceptibility. Results: Isolates were identified as L monocytogenes (259; 82.5%), L ivanovii (2; 0.6%), and Listeria species (53; 16.9%). Distribution of L monocytogenes serotypes: 4a/4c (0.4%), 4b (11.2%), 4d/4e (14%), 4b/4d/4e (9.3%), 1/2a (26.3%), 3a (7.7%), 1/2a/3a (6.2%), 1/2b/3b (1.2%), 1/2c (5%), 3c (1.2%), and 1/2c/3c (5.4%). Thirty-two L monocytogenes isolates (12.4%) were not typeable by PCR-REA, suggesting the possibility of serotypes 4ab/7. Susceptibility was 84.2% to 100% for most antimicrobials. Major resistance (R) and intermediate (I) susceptibility were found for clindamycin (R = 36.7%, I = 39.8% for L monocytogenes; R = 100% for L ivanovii; and R = 14%, I = 86% for Listeria species). Drugs of choice for treatment of human listeriosis (penicillin, ampicillin, and trimethoprim-sulfamethoxazole) remained effective; 1.2% of L monocytogenes were β-lactam resistant. Multidrug resistance was found only in L monocytogenes (26.6%) and Listeria species (26.4%), with (clindamycinI or R -erythromycinR-azithromycinR) and (ciprofloxacinI-clindamycinI) the most frequent phenotypes. Implications: Resistance to clindamycin and ciprofloxacin are shared between L monocytogenes and untyped Listeria. Although erythromycin is a drug of choice for prophylaxis in Colombian swine, resistance is low. No specific relationships between serotypes, sources, and antimicrobial susceptibility were found. | ResumenObjetivo: Analizar la distribución de serotipos de Listeria monocytogenes y la susceptibilidad antimicrobiana de los aislados de Listeria de plantas procesadoras de cerdos doméstica. Materiales y métodos: Los supuestos (314) aislados de Listeria fueron identificados molecularmente para distinguir entre las especies de L monocytogenes, Listeria ivanovii, y Listeria. Se identificaron los serotipos de Listeria monocytogenes a través de la reacción en cadena de la polimerasa (PCR por sus siglas en inglés) y del análisis de restricción de enzima (PCR-REA) y en busca de susceptibilidad antimicrobiana. Resultados: Los aislados fueron identificados como especies de L monocytogenes (259; 82.5%), L ivanovii (2; 0.6%), y Listeria (53; 16.9%). La distribución de serotipos de L monocytogenes: 4a/4c (0.4%), 4b (11.2%), 4d/4e (14%), 4b/4d/4e (9.3%), 1/2a (26.3%), 3a (7.7%), 1/2a/3a (6.2%), 1/2b/3b (1.2%), 1/2c (5%), 3c (1.2%), y 1/2c/3c (5.4%). Treinta y dos aislados de L monocytogenes (12.4%) no fueron tipificables a través de PCR-REA, sugiriendo la posibilidad de los serotipos 4ab/7. La susceptibilidad fue de 84.2% a 100% para la mayoría de los antimicrobianos. Se encontró mayor resistencia (R por sus siglas en inglés) y susceptibilidad intermedia (I por sus siglas en inglés) a la clindamicina (R = 36.7%, I = 39.8% para la especie L monocytogenes; R = 100% para la L ivanovii; y R = 14%, I = 86% para la Listeria). Las drogas de elección para el tratamiento de listeriosis humana (penicilina, ampicilina, y trimethoprim-sulfamethoxazole) permanecieron efectivas; 1.2% de L monocytogenes fueron resistentes a la β-lactam. La resistencia a fármacos múltiples se encontró sólo en las especies de L monocytogenes (26.6%) y Listeria (26.4%), con (clindamycinaI ó R-erytromicinaR-azitromicinaR) y (ciprofloxacinaI-clindamycinaI) siendo los fenotipos más frecuentes. | ResuméObjectif: Analyser la distribution des sérotypes de Listeria monocytogenes et la sensibilité antimicrobienne d’isolats de Listeria provenant d’une usine de transformation de porcs. Matériels et méthodes: Des isolats présumés de Listeria (314) ont été identifiés de façon moléculaire afin de distinguer L monocytogenes, Listeria ivanovii et Listeria sp. Les sérotypes de L monocytogenes ont été identifiés par réaction d’amplification en chaîne par la polymérase (PCR), PCR-analyse par enzyme de restriction (PCR-REA) et testés pour leur sensibilité aux antibiotiques. Résultats: Les isolats furent identifiés comme étant L monocytogenes (259; 82,5%), L ivanovii (2; 0,06%), et Listeria sp (53; 16,9%). La distribution des sérotypes de L monocytogenes était la suivante : 4a/4c (0,4%), 4b (11,2%), 4d/4e (14%), 4b/4d/4e (9,3%), 1/2a (26,3%), 3a (7,7%), 1/2a/3a (6,2%), 1/2b/3b (1,2%), 1/2c (5%), 3c (1,2%), et 1/2c/3c (5,4%). Trente-deux isolats de L monocytogenes (12,4%) étaient non-typables par PCR-REA, suggérant la possibilité de sérotypes 4a/7. La sensibilité envers la plupart des antimicrobiens variait de 84,2% à 100%. Les principales résistances (R) et sensibilités intermédiaires (I) ont été notées pour la clindamycine (R = 36,7%, I = 39,8% pour L monocytogenes; R = 100% pour L ivanovii; et R = 14%, I = 86% pour Listeria sp). Les antibiotiques de choix pour le traitement de la listériose humaine (pénicilline, ampicilline, et trimethoprime-sulfaméthoxazole) demeurent efficaces; 1,2% des isolats de L monocytogenes étaient résistants aux β-lactames. De la résistance multiple fut détectée chez L monocytogenes (26,6%) et Listeria sp (26,4%), les phénotypes les plus fréquents étant (clindamycineI ou R-érythromycineR-azithromycineR) et (ciprofloxacineI-clindamycineI). |
Keywords: swine, Listeria monocytogenes, Listeria ivanovii, antimicrobial susceptibility, molecular serotyping
Search the AASV web site
for pages with similar keywords.
Received: April 30, 2012
Accepted: June 29, 2012
The genus Listeria includes eight species,1 but only Listeria monocytogenes is a human and animal pathogen. Listeria ivanovii has long been considered a ruminant pathogen with a few reported cases of human infections.2-4 However, it has recently been proposed that L ivanovii is an opportunistic enteric human pathogen.5 Lack of central nervous system (CNS) involvement is probably a general characteristic of L ivanovii infection regardless of host species.6
Listeria monocytogenes infection in humans can affect the CNS, causing death or neurological sequelae. The infection may cause meningitis, meningoencephalitis, septicemia, abortion, and prenatal infection (invasive form of listeriosis), while the non-invasive form causes a gastrointestinal syndrome.7 In ruminants, listeriosis can cause four clinical syndromes: CNS infection causing meningoencephalitis in adults and meningitis in young animals; uterine infections characterized by abortion or neonatal septicemia; generalized septicemia with involvement of the liver and other organs; and mastitis in dairy cattle.8
In general, listeriosis affects at-risk groups such as infants, pregnant women,9 the elderly, and immunocompromised people,10 with mortality rate approximately 20% to 30% (within the risk groups).11 Occurrence of disease is related to consumption of contaminated food, principally ready-to-eat products, but in sporadic outbreaks and epidemics, a wide variety of foods act as vehicles: milk, cheese, paté, beef, pork, poultry meat, vegetables, and seafood.12,13
All strains of L monocytogenes are considered potentially pathogenic, but their virulence is variable. Serotypes frequently involved in development of food-borne listeriosis are 4b, 1/2b, 1/2c, and 1/2a;1 however, serotype 4b is considered the most virulent and is responsible for the majority of food-borne outbreaks and sporadic cases of illness.14-16 This may be related to the fact that this serotype is more adapted to the mammalian host than are the strains corresponding to serotype 1/2.7,17,18
In recent years, pork production in Colombia has increased, which has expanded participation in the domestic market.19-21 Clinical listeriosis rarely occurs in pigs. The primary manifestation of the disease in pigs is septicemia, with encephalitis and abortions occurring less commonly. However, infection with L monocytogenes is usually subclinical, ie, pigs are asymptomatic carriers,8 with a prevalence of 10% to 50% in fecal samples.22 It is difficult to find data on the economic impact of listeriosis on the swine industry. However, economics should not be the only motive for improving surveillance, detection methodologies, disinfection, and handling of this organism. It is also necessary to consider the role of animals as a reservoir and source of infection for humans. Human infection can occur by direct contact or through consumption of contaminated food from animal sources.8
Few published or accessible studies are available on antimicrobial susceptibility patterns of L monocytogenes in Colombia,23 and few publications relate classical or molecular serotyping to food type and source.24-26 A recent study27 showed that the prevalence of L monocytogenes in pig carcasses, pig-meat cuts, and pig derivates in Colombia was approximately 13.8%. A study conducted by the Secretaría Distrital de Salud de Bogotá, Colombia, between 2001 and 2004 reported an overall incidence of 11.2% and incidences of 6.3%, 2.0%, and 0.2% in hams, chorizo (a type of highly seasoned pork sausage), and other sausages, respectively.28
In this study we included Listeria isolates obtained from two concurrent studies performed at processing in Colombia, 2010-2011. These studies were conducted to investigate prevalence, antimicrobial susceptibility, disinfectant tolerance, and serotypes of L monocytogenes in circulation, with the main objective to analyze L monocytogenes serotype distribution and antimicrobial susceptibility of L monocytogenes, L ivanovii, and Listeria species in Colombia’s domestic pork industry.
Materials and methods
Sampling and processing
Sampling and processing were performed as described in Gamboa-Marín et al,27 except for surfaces, which were sampled and processed as described by the United States Department of Agriculture Food Safety and Inspection Service.29
Sources of isolates
Swine industry activity in Colombia is grouped into six regions according to the geographical distribution of technologically advanced hog farms, where most industrial activity is concentrated in the Antioquia, Western, and Central regions (Figure 1).21 A total of 314 presumptive Listeria species isolates from samples collected from swine processing facilities in the six pork-producing regions were used in this study.
Figure 1: National distribution of pork-industry production in Colombia.21  |
Origins of the isolates were distributed as follows: meat deboning (n = 172; 54.7%), pig carcasses (n = 26; 8.3%), ham (n = 13; 4.1%), sausage (n = 12; 3.8%), chorizo (n = 14; 4.5%), “longaniza” (another type of highly seasoned pork sausage; n = 19; 6.1%), utensils (n = 9; 2.9%), equipment (n = 25; 8%), contact surfaces (in contact with meat during processing ), (n = 8; 2.5%), non-contact surfaces (n = 13; 4.1%), hose water (n = 1; 0.3%), and worker stools (from workers who had diarrhea when other samples were collected; n = 2; 0.6%).
Genomic DNA purification and quantification
Presumptive isolates (314 isolates) were cultivated in brain heart infusion broth supplemented with 0.5% glucose (w/v) and incubated in a shaker for 12 hours at 37°C and 250 rpm.25 A 1-mL sample of culture was collected for DNA purification using the Wizard Genomic DNA Purification Kit (Promega, Fitchburg, Wisconsin). Purity and concentration of DNA were determined with a spectrophotometer (Biospec 1601; Shimadzu, Nakag Kyoto, Japan) (λ260/λ280 nm) with background correction set at λ320 nm.30
Molecular identification
Listeria monocytogenes and Listeria species identification. Two sets of primers, L1-Forward (CTC CAT AAA GGT GAC CCT) and U1-Reverse (CAG CMG CCG CGG TAA TWC), which yield a 938-bp product and identify the Listeria genus, and LF-Forward (CAA ACG TTA ACA ACG CAG TA) and LR-Reverse (TCC AGA GTG ATC GAT GTT AA), which yield a 750-bp product and amplify the hlyA gene typical of L monocytogenes, were employed in a multiplex-PCR to divide the isolates into two groups, L monocytogenes and Listeria species.25 The final reaction volume was 35 μL, composed of 1× PCR buffer, 1.5 mM of MgCl2, 0.2 mM of deoxyribonucleotides (dNTPs), 20 pmol of primers, and 2 U of Taq-deoxyribonucleic acid polymerase (TaqDNA Polymerase; Vivantis Technologies, Selangor Darul Ehsan, Malaysia). Five µL of DNA (approximately 100 ng) was used for amplification. Temperature settings were 95°C for 1 minute then 94°C for 30 seconds, followed by 40 cycles of 51°C for 20 seconds and 72°C for 30 seconds; then 72°C for 8 minutes.25
Listeria ivanovii identification. Another set of primers, IvaI-Forward (CTA CTC AAG CGC AAG CGG CAC) and Lis1B-Reverse (TTA TAC GCG ACC GAA GCC AAC), were used in a single PCR to amplify a 1100-bp fragment of L ivanovii. The final reaction volume was 25 µL, composed of 1× PCR buffer, 1.5 mM of MgCl2, 0.2 mM of dNTPs, 1×10-5 mg primers, and 1.5 U of TaqDNA Polymerase (Vivantis). Five µL of DNA (approximately 100 ng) was used for amplification. Temperature settings were 35 cycles of 95°C for 15 seconds, 62°C for 30 seconds, and 72°C for 50 seconds.31 The PCR product expected is approximately 1100 bp.
Grouping L monocytogenes isolates by divisions
The L monocytogenes isolates were sorted into divisions by use of a multiplex PCR. Two pairs of primers were employed: D1-Forward (CGA TAT TTT ATC TAC TTT GTC A), D1-Reverse (TTG CTC CAA AGC AGG GCA T), which yields a 214-bp product and classifies isolates as division I (serotypes 1/2b, 3b, 4b, 4d, and 4e) or division III (serotypes 4a and 4c), and D2 Forward (GCG GAG AAA GCT ATC GCA), Reverse (TTG TTC AAA CAT AGG GCT A), which yields a 140-bp product and classifies the isolates as division II (serotypes 1/2a, 1/2c, 3a, and 3c). Temperature settings were 95°C for 3 minutes followed by 25 cycles of 95°C for 30 seconds, 59°C for 30 seconds, and 72°C for 1 minute, then 72°C for 10 minutes.25
PCR serotyping
Isolates belonging to division II were subtyped using the FlaA primer set Forward (TTA CTA GAT CAA ACT GCT CC) and Reverse (AAG AAA AGC CCC TCG TCC), to generate a 538-bp product characteristic of serotypes 1/2a and 3a. Absence of amplification identified serotype 1/2c or 3c. Temperature settings were 95°C for 3 minutes followed by 25 cycles of 95°C for 30 seconds, 54°C for 30 seconds, and 72°C for 1 minute, then 72°C for 10 minutes.25
Isolates belonging to divisions I and III were subtyped using the GLT primer set Forward (AAA GTG AGT TCT TAC GAG ATT T) and Reverse (AAT TAG GAA ATC GAC CTT CT) to obtain a 483-bp product characteristic of serotypes 1/2b and 3b. Temperature settings were 95°C for 3 minutes followed by 25 cycles of 95°C for 30 seconds, 45°C for 30 seconds, and 72°C for 1 minute, then 72°C for 10 minutes.25
The reaction mixture for D1/D2, FlaA, and GLT consisted of 25 µL reaction volume, 50 pmol per µL of each primer, 1U of Taq DNA Polymerase (Vivantis), 1× PCR buffer, 0.2 mM of dNTPs, 2.5 mM of MgCl2, and 100 ng of DNA.25
Isolates that did not amplify a 483-bp band with the GLT primer set were assumed to be serotype 4 and thus were further subtyped with primers MAMA-C (LM4/LMB) Forward (CAG TTG CAA GCG CTT GGA GT) and Reverse (GTA AGT CTC CGA GGT TGC AA), yielding a 268-bp amplification product that identifies serotypes 4a and 4c.25 Strains that did not amplify were considered to be serotype 4 (b, d, or e). The reaction volume was 50 µL containing 0.5 µmol of each primer, 2U Taq DNA Polymerase (Vivantis), 1× PCR buffer, 0.2 mM of dNTPs, 2.0 mM of MgCl2, and 100 ng of DNA.25 MAMA-C PCR temperature settings were 95°C for 10 minutes followed by 40 cycles of 95°C for 30 seconds, 55°C for 1 minute, and 72°C for 1 minute, then 72°C for 10 minutes.
PCR-restriction enzyme analysis for confirmation of serotypes 1/2a, 1/2c, and 4b
PCR for iap gene amplification. Listeria monocytogenes isolates that were classified as serotypes 1/2a and 3a (identified as FlaA-positive by amplification and generation of a 538-bp product) and those that did not amplify with FlaA (serotype 1/2c or 3c), and the isolates that did not amplify with MAMA-C were considered to be serotype 4b, 4d, or 4e and were subtyped with a single PCR, using the primer set CLM1-Forward (ACA GCT GGG ATT GCG GT) and CLM2-Reverse (CCC AGC CAG AGC CGT GGA), located within the iap gene of L monocytogenes, in order to amplify a 1395-bp fragment of the iap gene. The reaction volume was 50 µL containing 0.1 pmol of each primer, 1.25 U Taq DNA Polymerase (Vivantis), 1× PCR buffer, 0.125 mM of dNTPs, 1.5 mM of MgCl2, and 100 ng of DNA. Temperature settings were 95°C for 5 minutes followed by 35 cycles of 95°C for 90 seconds, 54°C for 60 seconds, and 72°C for 3 minutes, then 72°C for 7 minutes.32
Restriction enzyme analysis. Five μL of each CLM1/CLM2 PCR product were digested with HindIII restriction enzyme (Vivantis) according to the instructions of the manufacturer.32 All PCR procedure temperatures were controlled in a C1000 Thermal Cycler (BioRad, Hercules, California). All PCR products and PCR-REA products were visualized in 1.5% agarose electrophoresis gel (w/v) prepared in TAE IX (40 mM Tris acetate, 1 mM EDTA, pH 8 ± 0.2) and stained with ethidium bromide (0.3 µg per mL) and run at 100 V in power supply model PowerPac Basic (BioRad) for 1 hour, then visualized and photographed under UV light. A 100-bp ladder (Promega or Axygen Biosciences, Union City, California) was used as a molecular size marker and L monocytogenes ATCC 1911533 was used as a PCR control.
Antimicrobial susceptibility test. A broth microdilution technique (MicroScan System; Siemens Healthcare Diagnostics, Bogotá, DC, Colombia) was employed for antimicrobial susceptibility testing of L monocytogenes and L ivanovii isolates. A cell suspension equivalent to 0.5 on the McFarland scale, prepared in Müeller-Hinton medium supplemented with lysed horse blood, was inoculated into the MICroSTREP plus 3 panel (Siemens) (Table 1). Panels were incubated following the manufacturer’s recommendations. The panel MICroSTREP plus 3 allowed fulfillment of the requirement stated in the M100-S22 document by Clinical and Laboratory Standards Institute (CLSI)34 concerning use of lysed horse blood for detection of antimicrobial susceptibility of exigent microorganisms. Streptococcus pneumoniae (ATCC 49619) was used as a control for antimicrobial susceptibility testing. Whonet 5.6 software (World Health Organization, 2010; http://www.whonet.org/dnn/Software/WHONET/tabid/97/language/en-US/Default.aspx ) was used for descriptive statistical analysis.23,25
Table 1: Antibiotics, terms, and their abbreviations in a study of Listeria species in samples collected from Colombian swine processing facilities
* MIC50 and MIC90 represent the MIC values of an antimicrobial at which the growth of 50% and 90% of the microbial population, respectively, are inhibited. † Isolates with MICs above or zone diameters below the value indicated for the susceptible break point are reported as nonsusceptible (NS) due to the absence or rare occurrence of resistant strains.34 |
Results
Molecular identification and serotyping
All isolates were confirmed by PCR to be Listeria species, ie, amplification of a 938-bp fragment with primers L1/U1. Of these, 259 isolates were identified as L monocytogenes (82.5%), ie, amplification of a 750-bp fragment with primers LF/LR. Only two isolates were identified as L ivanovii (0.6%) (data not shown), ie, amplification of a 1100-bp fragment with primers IvaI/Lis1B. The other 53 isolates were classified as Listeria species (16.9%) because DNA of these isolates amplified only with primers L1/U1. The control strain (L monocytogenes ATCC 19115) showed the expected bands (Figure 2A, B, C, and D).
Figure 2: Molecular identification and serotyping of Listeria isolates from samples collected in Colombian swine processing facilities. Panel A. 1, 9, and 16: 100-bp molecular size marker (Invitrogene); 2: reagent control for polymerase chain reaction (PCR); 3: DNA from L monocytogenes ATCC 19115 amplified with U1/L1-LF-LR; 4: DNA from Listeria species isolate amplified with U1-L1/LF-LR ( 938- and 750-bp bands identify L monocytogenes); 5 and 6: DNA from Listeria species amplified with U1/L1-LF-LR (938-bp band identifies Listeria genus); 7: DNA from L monocytogenes ATCC 19115 amplified with D1; 8: DNA from L monocytogenes isolates amplified with D1 (214-bp band identifies division I or III isolates); 10 and 11: DNA from L monocytogenes isolates amplified with D2 (140-bp band identifies division II isolates); 12 and 13: DNA from L monocytogenes isolates that do not amplify with D1/D2; 14 and 15: DNA from L monocytogenes isolates amplified with FlaA (538-bp band identifies serotypes 1/2a and 3a). Panel B. 1 and 9: 100-bp molecular size marker (Promega); 2 and 3: DNA from L monocytogenes isolates amplified with FlaA (538-bp band identifies serotypes 1/2a and 3a); 4, 5, and 7: DNA from L monocytogenes isolates amplified with GLT (483-bp band identifies serotypes 1/2b and 3b); 6: DNA from L monocytogenes isolates amplified with GLT; 8: Reagent control for PCR reaction. Panel C. 5: 100-bp molecular size marker (Axygen); 1, 3, 4, 7, and 8: DNA from L monocytogenes isolates amplified with CLM1-CLM2 (1395-bp band identifies the iap gene); 2, 6, and 9: empty wells. Panel D. 5: 100-bp molecular size marker (Axygen); 1, 2, 3, and 4: restriction enzyme analysis (REA) (HindIII) of DNA from L monocytogenes isolates after amplification with CLM1-CLM2 (693-, 425-, and 277-bp bands identifies serotypes 1/2a or 1/2c). Panel E. 1: 100-bp molecular size marker (Axygen); 2-4, 6, and 7: REA (HindIII) of DNA from L monocytogenes isolates after amplification with CLM1-CLM2 (1118- and 277-bp bands identifies serotype 4b); 5: negative REA (HindIII) of DNA from L monocytogenes isolates (no bands identifies serotypes 4d, 4e). 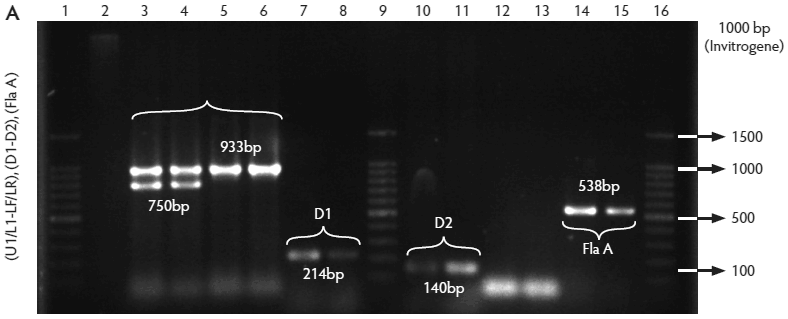 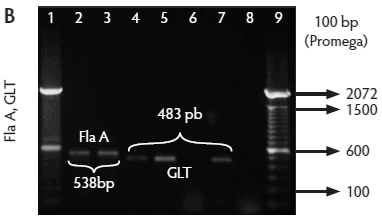 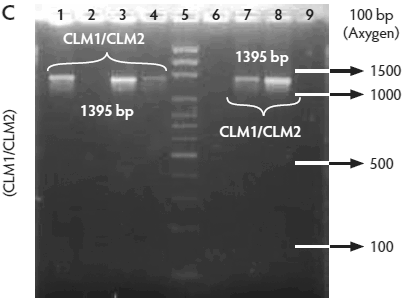 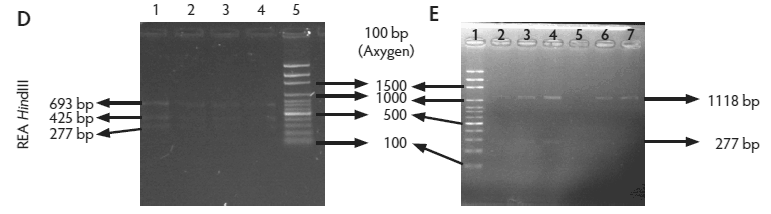 |
Molecular serotyping of L monocytogenes allowed detection of the following serotypes: 4a/4c (0.4%), 4b (11.2%), 4d/4e (14%), 4b/4d/4e (9.3%), 1/2a (26.3%), 3a (7.7%), 1/2a/3a (6.2%), 1/2b/3b (1.2%), 1/2c (5%), 3c (1.2%), and 1/2c/3c (5.4%). Thirty-two L monocytogenes isolates (12.4%) were not serotypes identified by the specific primers, suggesting the possibility that they were serotypes 4ab or 7, but this remains to be demonstrated. The control strain (L monocytogenes ATCC 19115) was serotyped as 1/2b as expected.
Antimicrobial susceptibility test
The antimicrobial susceptibility, resistance, and intermediate patterns of the 314 isolates can be seen in Tables 2, 3, and 4. In this study, the susceptibility of isolates varied between 84.2% and 100% for most antimicrobials. Major resistance and intermediate values were found for clindamycin: R = 36.7%, I = 39.8% for L monocytogenes (Table 2); R = 100% for L ivanovii (Table 3); and R=14%, I=86% for Listeria species (Table 4). The primary drugs of choice against listeriosis (penicillin, ampicillin, and trimethoprim-sulfamethoxazole) remained effective for most isolates, and only 1.2% of L monocytogenes isolates were nonsusceptible to penicillin and ampicillin.
Table 2: Number and the distribution of 259 Listeria monocytogenes isolates collected from Colombian swine processing facilities in terms of susceptibility to antibiotics commonly used in humans*
* Antimicrobial abbreviations and break-point categories are defined in Table 1. Bold print identifies the primary drugs of choice for treatment of human listeriosis and the greatest numbers of organisms in the R and I susceptibility categories. † Break points for Staphylococcus species used.23 ‡ Break points for Enterococcus species used.23 MIC = minimal inhibitory concentration; NA = not applicable (break points not yet established by the Clinical and Laboratory Standards Institute). |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Table 3: Number and distribution of two Listeria ivanovii isolates collected from Colombian swine processing facilities in terms of susceptibility to antibiotics commonly used in humans*
* Antimicrobial abbreviations and break-point categories are defined in Table 1. Bold print identifies the primary drugs of choice for treatment of human listeriosis and the greatest numbers of organisms in the R and I susceptibility categories. † Break points for Staphylococcus species used.23 ‡ Break points for Enterococcus species used.23 MIC = minimal inhibitory concentration. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Table 4: Number and distribution of 53 Listeria species isolates collected from Colombian swine processing facilities in terms of susceptibility to antibiotics commonly used in humans*
* Antimicrobial abbreviations and break-point categories are defined in Table 1. Bold print identifies the primary drugs of choice for treatment of human listeriosis and the greatest numbers of organisms in the R and I susceptibility categories. † Breakpoints for Staphylococcus species used.23 ‡ Break points for Enterococcus species used.23 MIC = minimal inhibitory concentration |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
On the basis of criteria established by the CLSI34 for Staphylococcus species and Enterococcus species, 26.6% of L monocytogenes and 26.4% of Listeria species displayed multidrug resistance. A total of 30 multidrug-resistance patterns were identified in L monocytogenes isolates, whereas in Listeria species, only three patterns were identified. The most frequent phenotypes for L monocytogenes were clindamycinI-erythromycinR-azithromycin,R and clindamycinR-erythromycinR-azithromycin,R while for Listeria species, the most frequent phenotype was ciprofloxacinI-clindamycin.I No multidrug-resistance was found in L ivanovii. It is important to note that 19 of 69 L monocytogenes multidrug-resistant isolates (27.5%) showed simultaneous resistance to clindamycin and erythromycin (Table 5); no chloramphenicolR phenotype was identified among these isolates. This combination was not found in any L ivanovii strains or in Listeria species (Table 6).
Table 5: Distribution of Listeria monocytogenes MDR isolates in samples collected from Colombian swine processing facilities*
* Antimicrobial abbreviations and break-point categories are defined in Table 1. † MDR patterns occurred when MIC values for an isolate classified it in the I or R susceptibility category for two or more classes of antimicrobials, regardless of intrinsic resistance to cephalosporins. MDR = multidrug-resistant; MIC = minimal inhibitory concentration. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Table 6: Distribution of Listeria species MDR isolates in samples collected from Colombian swine processing facilities*
* Antimicrobial abbreviations and break-point categories are defined in Table 1. † Highly seasoned specialty sausages. MDR = multidrug-resistant. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
The results of antimicrobial susceptibility testing for cefotaxime, cefepime, cephradine, and cefuroxime were not reported in this paper. However it is important to note that resistance to cephalosporins was confirmed in more than 90% of isolates (data not shown).
Relationship between serotype, source, and antimicrobial resistance
The relationship between antimicrobial resistance (including intermediate isolates), L monocytogenes serotypes, and distribution by source revealed that, first, major resistance behavior was found against clindamycin. Second, major diversity of resistance patterns was found in serotypes 4b (17 patterns), 4b/4d/4e (15 patterns), 1/2c/3c (11 patterns), and 3a (nine patterns). Third, serotype 1/2b/3b was found uniquely in equipment, whereas only one isolate (serotype 4a/4c) was found on a contact surface. Fourth, the other serotypes (4b, 4d/4e, and 1/2a/3a) were found in all sources.
The most complex resistance phenotypes were found in isolates of serotypes 1/2a/3a and 1/2c/3c (with resistance to up to five antimicrobial agents plus intrinsic resistance to cephalosporin). It should be emphasized that presumptive isolates of serotype 4ab or 7 (to be confirmed) also showed an important antimicrobial resistance pattern.
Discussion
In a recent study, we reported an overall prevalence of L monocytogenes of approximately 13.8% from Colombian swine processing plants,28 but prevalence alone should not be considered. Antimicrobial susceptibility in association with serotype distribution will provide important information for health and food agencies that must evaluate and control the microbiological risk in a population.
In Colombia, there are few published data or easily accessible documents concerning the presence and distribution of L monocytogenes serotypes.25 It is well known1 that certain serotypes occur more often in food or in infected humans or animals, and it is also known that approximately 95% of strains isolated from cases of human listeriosis caused by L monocytogenes correspond mainly to four serotypes: 4b, 1/2b, 1/2c, and 1/2a. In two previous studies we used molecular methods to determine the serotypes of L monocytogenes isolates from different sources and types of food, and we then analyzed the antimicrobial susceptibility patterns of these isolates.23,25 Herein we have started with a representative sample of the swine processing plants,27 in order to analyze the antimicrobial susceptibility and serotype distribution along the pork-meat chain.
We combined the molecular serotyping methodology used in our earlier study25 with a HindIII restriction enzyme analysis after iap gene amplification,32 with the main objective of discriminating among L monocytogenes serotypes 1/2a, 1/2c, and 4b (Figure 2). There is an important discrimination to be made between L ivanovii and L monocytogenes, as L monocytogenes is a human and animal pathogen, while L ivanovii is a ruminant pathogen that has recently been recognized as an opportunistic human enteric pathogen.5
Our results show a high frequency of serotypes 1/2a (26.3%) followed by 4d/4e (14%), 4b (11.2%), and 4b/4d/4e (9.3%). These frequencies are lower than those obtained by other authors (64%,35 57%,36 and 92.8%,37 respectively); nonetheless, our results are in agreement that serotype 1/2a is the most frequent. The frequency of serotype 4b in this study is higher than reported by other authors,38 who investigated the occurrence of L monocytogenes in sausages and found serovar 4b in 8.0% of isolates and the highest prevalence in serotype 1/2a (49.5%).
The presence and persistence of L monocytogenes lineage II isolates (serotypes 1/2a, 1/2c, 3a, and 3c) in food and food-plant environments may be associated with a greater capacity for growth and survival.1 Strains of lineage II may be more competent than those belonging to lineage I (serotypes 1/2b, 3b, 4b, 4d, and 4e), which probably is associated with increased resistance to bacteriocins, although, after storage at 4°C, serotype 4b strains tend to be more resistant to heat treatment (60°C) than 1/2a strains.1
Previous studies11,39,40 show that the strains of serotype 1/2 presented a higher food prevalence; nonetheless, serotype 4b is most often isolated from patients with listeriosis, suggesting that this serotype is more pathogenic than the others.1 Additionally, serotype 4b is more adapted to the human host.17,18
On the one hand, some investigations11,17,18,39,40 clearly show that serotype 1/2 has been isolated from various food products, suggesting that it has different ecological niches. On the other hand, our study found that serotypes 1/2a, 3a, and 4b/4d/4e were detected in all types of samples, which is important, considering that serotypes 1/2a and 4b have been the causes of several listeriosis outbreaks,41 including those associated with consumption of pork.
Considering that L monocytogenes is resistant to 3rd to 6th generation cephalosporins, that these antimicrobials are included in the MICroSTREP plus3 panel, and that cephalosporin resistance has been reported before,41 patterns of cephalosporin resistance were not included in the multidrug resistance analysis and will not be further discussed here.
In a previous study,23 we assayed the antimicrobial susceptibility of L monocytogenes isolates from several sources, finding major resistance (30% to 65% of isolates) to clindamycin, meropenem, rifampin, and ciprofloxacin, whereas penicillin, ampicillin, and trimethoprim-sulfamethoxazole, the primary drugs of choice against listeriosis,23 remained effective for most isolates (84%). As in the previous study,23 clindamycin resistance was the most common resistant phenotype (approximately 80.6% of phenotypes) among all Listeria isolates, while resistance to meropenem, rifampin, and ciprofloxacin comprised 0.4% to 24.5% of resistant phenotypes. Results of the present study do not agree with the recently reported decrease in susceptibility of L monocytogenes to ciprofloxacin.42
All Listeria isolates studied displayed approximately 98% susceptibility to β-lactams, with only 1.2% of isolates classified as nonsusceptible. In our previous study,23 16% were classified as nonsusceptible.
More than 90% of isolates were susceptible to the antimicrobials assayed, with susceptibility to rifampin and vancomycin particularly notable because of their importance in treating Mycobacterium tuberculosis and Staphylococcus aureus infections, respectively.23
A phenomenon of inducible clindamycin resistance has been described in clinical isolates of Staphylococcus species, when an infection caused by an erythromycinR-clindamycinI or an erythromycinR-clindamycinS strain is being treated with clindamycin.23,34 It is not possible yet to extrapolate this phenomenon to Listeria species, although it is important to note that in the present study, 19 of 69 L monocytogenes isolates (27.5%) displayed both clindamycin and erythromycin resistance, suggesting a possible modification of the 23S rRNA.43 These values are higher than in our earlier reports of 7%23 and 10%25 of L monocytogenes isolates resistant to clindamycin and erythromycin, respectively. In some countries (eg, Cuba44 and Colombia45), erythromycin is commonly employed as a prophylactic antimicrobial in domestic pig herds.
In Listeria species, the multidrug-resistance patterns were limited to a combination of clindamycin-intermediate and ciprofloxacin-intermediate or resistant, but in L monocytogenes, 30 different patterns were detected. Most multidrug-resistant isolates (61.6%) were L monocytogenes isolated from pig carcasses, and the greatest number of multidrug-resistance patterns was found in deboned meat, with clindamycin, erythromycin, and azithromycin involved in most of the patterns. Resistance phenotypes reported in the present study were similar to those in the previous study,23 in which 78% of resistance phenotypes involved clindamycin, erythromycin, azithromycin, tetracycline, ciprofloxacin, rifampin, or meropenem. However, in the present study, 22% of resistance phenotypes involved trimethoprim-sulfamethoxazole and chloramphenicol.
Relationships between the source, the organism, and resistance to specific antimicrobials were not found. However, all serotypes were distributed among all types of samples, as expected from the operational dynamics of the swine processing plants. Isolates belonging to L monocytogenes serotypes 4, 1/2a, 3a, and 1/2c/3c showed more varied resistance patterns.
The geographic sources of serotypes and resistant, intermediate, or multidrug-resistant isolates will remain confidential. However, serotypes and antimicrobial susceptibility patterns found in the swine processing facilities or in specific sources in those facilities will be privately revealed to the industry authorities so they can take action.
In conclusion, various L monocytogenes and Listeria species strains have circulated in the domestic pork industry, and the multidrug-resistance phenotypes identified in this study showed 77% similarity to those detected by Ruiz-Bolivar et al.23 Considering the period of sampling and the isolate sources (food type and geographical region), this suggests “a stable spread of resistance patterns among L monocytogenes circulating in the country.”23 In contrast, it appears that these genes have not been disseminated to the Listeria species isolates, because in this study, only two resistance phenotypes (clindamycin and ciprofloxacin; 22%) were shared between L monocytogenes and Listeria species. It is not yet possible to associate a specific serotype with an antimicrobial resistance pattern. It seems that antimicrobial susceptibility related to the circulating strains of L monocytogenes has been stable over the past 5 years, at least in the studied areas. Further studies are required to determine relationships between isolates obtained from food, animals, and humans, and to extend the studies to other production chains.
Implications
• Under the conditions of this study, only two resistance phenotypes (clindamycin and ciprofloxacin) are shared between L monocytogenes and untyped Listeria.
• Under the conditions of this study, resistance is low to erythromycin, a drug of choice for prophylaxis in the domestic Colombian pork industry.
• Under the conditions of this study, specific relationships between serotypes, sources of isolates, and antimicrobial susceptibility are not detectable.
Acknowledgments
This work was supported by the Ministerio de Agricultura y Desarrollo Rural, Colombia through a research project (Grant 2008W4845) and Pontificia Universidad Javeriana, Bogotá, DC, Colombia (Grant 054 -2008W4497–3592). Finally the authors thank Ms Katty Díaz for English editing of the manuscript.
References
1. Orsi RH, den Bakker HC, Wiedmann M. Listeria monocytogenes lineages: Genomics, evolution, ecology, and phenotypic characteristics. Int J Med Microbiol. 2011;301:79–96.
2. Elischerova K, Cupkova E, Urgeova E, Lysy J, Sesevickova A. Isolation of Listeria ivanovii in Slovakia. Ceskoslovenska Epidemiologie, Mikrobiologie, Imunologie (Praha). 1990;39:228–236.
3. Snapir YM, Vaisbein E, Nassar F. Low virulence but potentially fatal outcome Listeria ivanovii. Eur J Int Med. 2006;17:286–287.
4. Cummings AJ, Fielding AK, McLaughlin J. Listeria ivanovii infection in a patient with AIDS. J Infect. 1994;28:89–91.
5. Guillet C, Join-Lambert O, Le Monnier A, Leclercq A, Mechaï F, Mamzer-Bruneel M-F, Bielecka MK, Scortti M, Disson O, Berche P, Vazquez-Boland J, Lortholary O, Lecuit M. Human listeriosis caused by Listeria ivanovii. Emerg Infect Dis. 2010;16:136–138.
6. Surve N, Bagde U. Toxicity of oxazolidinone linezolid on pathogenic microorganism Listeria ivanovii. Int J Biol. 2011;3:72–78. doi:10.5539/ijb.v3n4p72.
7. Torres KJ, Sierra SC, Poutou RA, Carrascal AK, Mercado M. Patogénesis de Listeria monocytogenes, microorganismo zoonótico emergente [Pathogenesis of Listeria monocytogenes, zoonotic emergent microorganism]. Revista MVZ-Córdoba. 2005;10:511–543.
8. Belalcazar ME, Poutou RA, Torres KJ, Gallegos JM, Torres O, Carrascal AK. Listeria monocytogenes y listeriosis animal [Listeria monocytogenes and animal listeriosis]. Revista UDCA Atualidad y Divulgación Científica. 2005;8:3–16.
9. Salazar CC, Cunha JSL, Schlatter D. Abortamento de repetiçao. Femina. 2001;29:667–672.
10. Huang YT, Chen SU, Wu MZ, Chen CY, Hsieh WS, Tsao BN, Horng CJ, Hsueh PR. Molecular evidence for vertical transmission of listeriosis, Taiwan. J Med Microbiol. 2006;55:1601–1603.
11. Todd ECD, Notermans S. Surveillance of listeriosis and its causative pathogen, Listeria monocytogenes. Food Cont. 2011;22:1484–1490.
12. Ballesteros L, Moreno Y, Cuesta G, Rodrigo A, Tomás D, Hernández M, Ferrús MA, García Henández J. Persistence of Listeria monocytogenes strains in a frozen vegetables processing plant determined by serotyping and REP-PCR. Int J Food Sci Technol. 2011;46:1109–1112.
13. Albarracín FY, Poutou RA, Carrascal AK. Listeria species, y L monocytogenes en leche cruda de cabra [Listeria species and Listeria monocytogenes in raw goat’s milk]. Revista MVZ-Córdoba. 2008;13:1326–1332.
14. Liu D, Lawrence ML, Gorski L, Mandrell RE, Ainsworth AJ, Austin FW. Listeria monocytogenes serotype 4b strains belonging to lineages I and III possess distinct molecular features. J Clin Microbiol. 2006;44:214–217.
15. Liu D. Identification, subtyping and virulence determination of Listeria monocytogenes, an important foodborne pathogen. J Med Microbiol. 2006;55:645–659.
16. Liu D, Lawrence ML, Wiedmann M, Gorski L, Mandrell RE, Ainsworth AJ, Austin FW. Listeria monocytogenes subgroups IIIA, IIIB, and IIIC delineate genetically distinct populations with varied pathogenic potential. J Clin Microbiol. 2006;44:4229–4233.
17. Vázquez J, Domínguez G, Gónzalez B, Kreft J, Goebel W. Pathogenicity islands and virulence evolution in Listeria. Microb Infect. 2001;3:571–584.
18. Vázquez J, Kuhn M, Berche P, Chakraborty T, Domínguez G, Goebel W, González B, Wehland J, Kreft J. Listeria pathogenesis and molecular virulence determinants. Clin Microbiol Rev. 2001;14:584–640.
19. Food and Agriculture Organization of the United Nations (FAOSTAT/FAO). Producción de carne de cerdo en Colombia y a nivel Mundial. 2009. Available at: http://faostat.fao.org/site/569/DesktopDefault.aspx?PageID=569#ancor. Accessed 6 October 2012.
20. Rosero O, Lukesová D. Food and perspectives on pig production system in Colombia. Agricult Trop Subtrop. 2008;41:122–127.
21. DANE. Sacrificio de ganado total nacional y regional - vacunos, porcinos y otras especies [Slaughtering regional and national total - cattle, pigs and other species]. 2010. Available at: http://www.dane.gov.co/en/index.php?option=com_content&view=article&id=175&Itemid=28. Accessed 26 August 2012.
22. Meng J, Doyle MP. Emerging issues in microbial food safety. Ann Rev Nut. 1997;17:255–275.
23. Ruiz-Bolivar Z, Neuque-Rico MC, Poutou-Piñales RA, Carrascal-Camacho AK, Máttar-Velilla S. Antimicrobial susceptibility of L monocytogenes food isolates from different cities of Colombia. Foodborne Path Dis. 2011;8:913–919.
24. Muñoz AI, Vargas M, Otero L, Díaz G, Guzmán V. Presencia de Listeria monocytogenes en alimentos listos para el consumo, procedentes de plazas de mercado y delicatessen de supermercados de cadena en Bogotá D.C. 2002-2008 [Presence of Listeria monocytogenes in ready-to-eat foods available in open markets, delicatessens and supermarkets, Bogotá, 2002-2008]. Biomédica. 2011;31:428–439.
25. Ruiz-Bolivar Z, Carrascal-Camacho AK, Neuque-Rico MC, Gutiérrez-Triviño C, Rodríguez-Bocanegra MX, Poutou-Piñales RA, Mattar S. Enterobacterial repetitive intergenic consensus-polymerase chain reaction (ERIC-PCR) fingerprinting reveals intra-serotype variations among circulating Listeria monocytogenes strains. Afr J Microbiol Res. 2011;5:1586–1598.
26. Medrano MV, Restrepo S, Vanegas MC. Tipificación molecular de Listeria monocytogenes aisaldas de muestras clínicas y alimentos. Biomédica. 2006;26:442–450.
27. Gamboa-Marín A, Buitrago SM, Pérez-Pérez KI, Mercado M, Poutou-Piñales RA, Carrascal-Camacho AK. Prevalence of Listeria monocytogenes in pork-meat and other processed products from the Colombian swine industry. Rev MVZ-Córdoba. 2012;17:2827–2833.
28. Vera H, Ferro CJ, Triana LM. 2006. Prevalencia de Listeria moncytogenes en derivados cárnicos cocidos para consumo directo analizados en el laboratorio de salud pública [Prevalence of Listeria moncytogenes in cooked meat products for direct consumption, analyzed in the public health laboratory]. Bogotá 1 de septiembre 2001 - 31 agosto de 2004. Available at: http://www.docstoc.com/docs/3180734/T%C3%8DTULO-PREVALENCIA-DE-LISTERIA-COCIDOS-PARA-MONOCYTOGENES-DIRECTO-EN-DERIVADOS. Accessed 4 October 2012.
29. USDA-FSIS. Isolation and Identification of Listeria monocytogenes from Red Meat, Poultry, Egg, and Environmental Samples. MLG 8.07. In: USDA/FSIS Microbiology Laboratory Guidebook. Washington, DC: United States Department of Agriculture Food Safety and Inspection Service; 2002.
30. Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3th ed. Cold Spring Harbour, New York: Cold Spring Harbor Laboratory Press; 2001.
31. Bubert A, Hein I, Rauch M, Lehner A, Yoon B, Goebel W, Wagner M. Detection and differentiation of Listeria species by a single reaction based on multiplex PCR. Appl Environ Microbiol. 1999;65:4688–4692.
32. Comi G, Cocolin L, Cantoni C, Manzano M. A RE-PCR method to distinguish Listeria monocytogenes serovars. FEMS Immunol Med Microbiol. 1997;18:99–104.
33. Gallegos JM, Vanegas MC, Albarracín Y, Máttar S, Poutou RA, Carrascal AK. Frequency of isolation of Listeria species, in different retail foods in Colombia. Anim Prod Res Adv. 2008;4:9–18.
34. CLSI. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Second Informational Supplement. M100-S22. Wayne, Pennsylvania: Clinical and Laboratory Standards Institute; 2012.
35. Chasseignaux E, Toquin MT, Ragimbeau C, Salvat G, Colin P, Ermel G. Molecular epidemiology of Listeria monocytogenes isolates collected from the environment, raw meat and raw products in two poultry and pork processing plants. J Appl Microbiol. 2001;91:888–899.
36. Jemmi T, Pak SI, Salman MD. Prevalence and risk factors for contamination with Listeria monocytogenes of imported and exported meat and fish products in Switzerland, 1992-2000. Prev Vet Med. 2002;54:25–36.
37. Vitas AI, Aguado V, Garcia-Jalon I. Occurrence of Listeria monocytogenes in fresh and processed foods in Navarra (Spain). Int J Food Microbiol. 2004; 90:349–356.
38. Thévenot D, Dernburg A, Vernozy-Rozand C. An updated review of Listeria monocytogenes in the pork meat industry and its products. J Appl Microbiol. 2006;101:7–17.
39. Ochiai Y, Yamada F, Batmunkh O, Mochizuki M, Takano T, Hondo R, Ueda F. Prevalence of Listeria monocytogenes in retailed meat in the Tokyo metropolitan area. J Food Prot. 2010;73:1688–1693.
40. Taillefer C, Boucher M, Laferrière C, Morin L. Perinatal listeriosis: Canada’s 2008 outbreaks. J Obstet Gynecol Canada. 2010;32:45–48.
41. Troxler R, Von Graevenitz A, Funke G, Wiedemann B, Stock I. Natural antibiotic susceptibility of Listeria species: L grayi, L innocua, L ivanovii, L monocytogenes, L seeligeri and L welshimeri strains. Clin Microbiol Infect. 2000;6:525–535.
42. De Nes F, Pelicioli Riboldi G, Guedes Frazzon AP, Alves d’Azevedo P, Frazzon J. Antimicrobial resistance and investigation of the molecular epidemiology of Listeria monocytogenes in dairy products. Rev Soc Bras Med Trop. 2010;43:382–385.
43. Davis JA, Jackson CR. Comparative antimicrobial susceptibility of Listeria monocytogenes, L innocua, and L welshimeri. Microb Drug Resist. 2009;15:27–32.
44. Lozano MC, Arias DC. Residuos de fármacos en alimentos de origen animal: panorama actual en Colombia [Drugs residues in foods of animal origin: actual prospect in Colombia]. Revista Colombiana de Ciencias Pecuarias. 2008;21:121–135.
45. Baez M, Espinosa I, Vichi J, Martínez S. Estudio de la sensibilidad in vitro frente a diferentes antimicrobianos en cepas de S. suis asociados a neumonía porcina [Study of in vitro sensitivity against different antimicrobials in S. suis strains associated with porcine pneumonia]. Revista Salud Animal. 2012;34:57–62.
